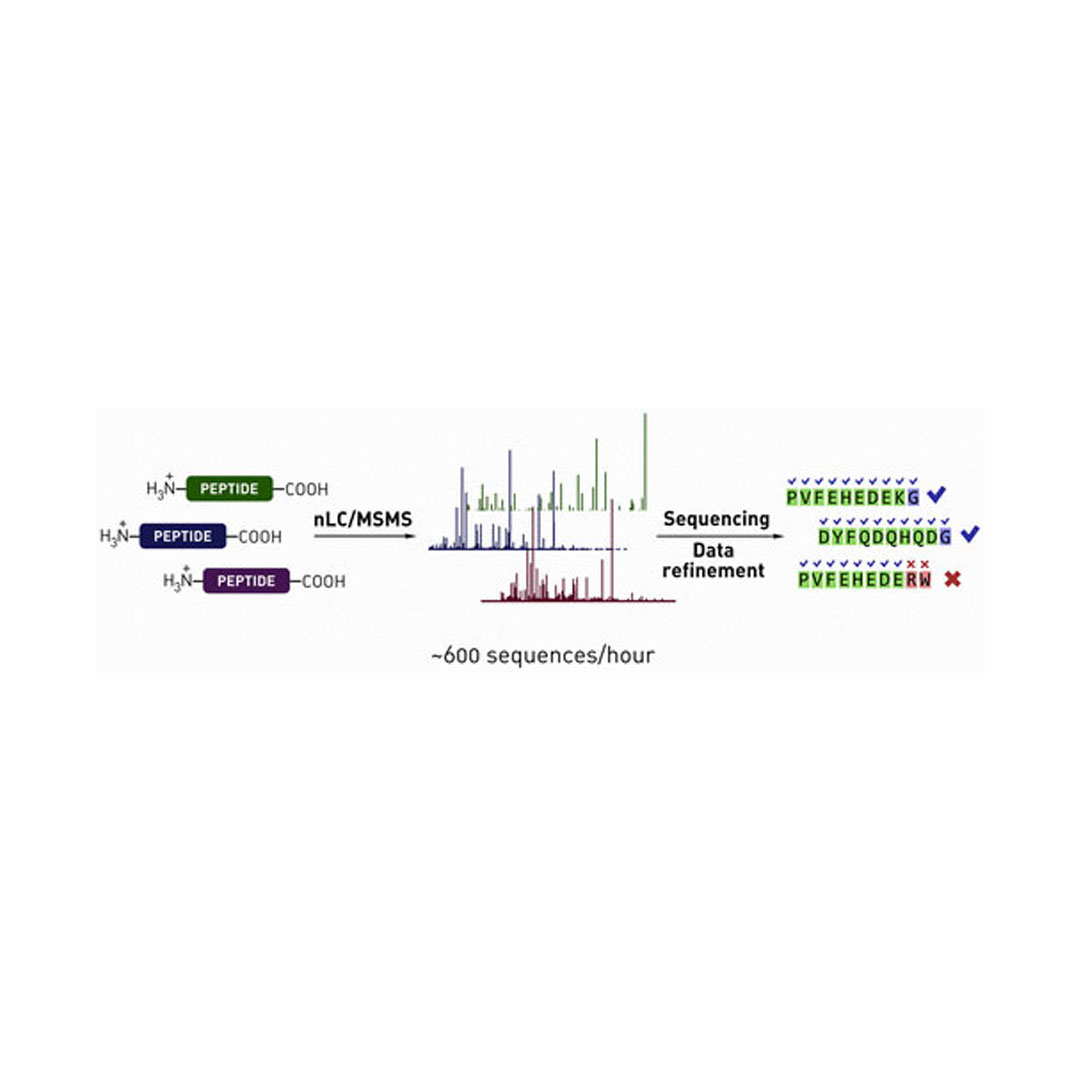
Library Design-Facilitated High-Throughput Sequencing of Synthetic Peptide Libraries
A methodology to achieve high-throughput de novo sequencing of synthetic peptide mixtures is reported. The approach leverages shotgun nanoliquid chromatography coupled with tandem mass spectrometry-based de novo sequencing of library mixtures (up to 2000 peptides) as well as automated data analysis protocols to filter away incorrect assignments, noise, and synthetic side-products. For increasing the confidence in the sequencing results, mass spectrometry-friendly library designs were developed that enabled unambiguous decoding of up to 600 peptide sequences per hour while maintaining greater than 85% sequence identification rates in most cases. The reliability of the reported decoding strategy was additionally confirmed by matching fragmentation spectra for select authentic peptides identified from library sequencing samples. The methods reported here are directly applicable to screening techniques that yield mixtures of active compounds, including particle sorting of one-bead one-compound libraries and affinity enrichment of synthetic library mixtures performed in solution.
Pentelute Lab, MIT, Cambridge, Chemistry, Molecular biology, technology development, peptide, protein-based therapeutics, chemical Biology, de novo sequencing, one-bead one-compound libraries, shotgun nanoliquid chromatography, synthetic peptide mixtures, tandem mass spectrometry,
17069
portfolio_page-template-default,single,single-portfolio_page,postid-17069,bridge-core-3.0.1,qode-page-transition-enabled,ajax_fade,page_not_loaded,,paspartu_enabled,paspartu_on_top_fixed,paspartu_on_bottom_fixed,qode_grid_1200,qode_popup_menu_push_text_top,qode-theme-ver-28.7,qode-theme-bridge,disabled_footer_top,wpb-js-composer js-comp-ver-6.8.0,vc_responsive
Library Design-Facilitated High-Throughput Sequencing of Synthetic Peptide Libraries
Library Design-Facilitated High-Throughput Sequencing of Synthetic Peptide Libraries
ACS Comb. Sci., 2017, 19 (11), pp 694–701
DOI: 10.1021/acscombsci.7b00109
Publication Date (Web): September 11, 2017
Alexander A. Vinogradov, Zachary P. Gates, Chi Zhang, Anthony J. Quartararo, Kathryn H. Halloran, and Bradley L. Pentelute
Abstract
A methodology to achieve high-throughput de novo sequencing of synthetic peptide mixtures is reported. The approach leverages shotgun nanoliquid chromatography coupled with tandem mass spectrometry-based de novo sequencing of library mixtures (up to 2000 peptides) as well as automated data analysis protocols to filter away incorrect assignments, noise, and synthetic side-products. For increasing the confidence in the sequencing results, mass spectrometry-friendly library designs were developed that enabled unambiguous decoding of up to 600 peptide sequences per hour while maintaining greater than 85% sequence identification rates in most cases. The reliability of the reported decoding strategy was additionally confirmed by matching fragmentation spectra for select authentic peptides identified from library sequencing samples. The methods reported here are directly applicable to screening techniques that yield mixtures of active compounds, including particle sorting of one-bead one-compound libraries and affinity enrichment of synthetic library mixtures performed in solution.
Category
2017, Publications